From Chaos to Clarity: The Apple TCP Genes Naming Makeover
Abstract
Apple Proliferation (AP) is a widespread disease affecting apple orchards throughout Europe, including South Tyrol.The disease is associated with a phytoplasma, 'Candidatus Phytoplasma mali' ('Ca. P. mali'), which manipulates the plant's physiological processes through the secretion of small peptides called effectors. One well-studied phytoplasma effector is SAP11, which targets Teosinte Branched 1/Cycloidea/Proliferating Cell Factor 1 (TCP) genes in multiple plant species, including apple. TCP genes encode plant-specific transcription factors involved in various biological processes, such as growth, development, and responses to stimuli. The identification and naming of TCP genes have been inconsistent, leading to confusion and redundancy in sequence databases. In apple, 52 TCP genes (MdTCP) were identified in 2014 on the 2010 assembly of the apple genome, which is now considered outdated. To address this issue, a comprehensive investigation was conducted to identify and name TCP genes in apple using the high-quality genome assembly, GDDH13v1.1. Existing TCP sequences were aligned with the genome excluding redundant, fragmented, and non-TCP sequences. The revised set comprised 40 unique MdTCPs, including three novel genes. To establish a standardised nomenclature, each MdTCP was BLASTedagainst the Arabidopsis thaliana gene database, in order to identify the best hits for each MdTCP. The MdTCP genes were then renamed, incorporating the letter "a" or "b" to differentiate between MdTCPs showing homology to the same AtTCP, and also between the existing and proposed nomenclatures. The present study highlights the need for clarity and organisation in sequence databases, especially in respect of TCP genes. The presence of redundant and fragmented sequences in databases complicates accurate identification and annotation of genes. By providing a comprehensive and standardised nomenclature system, we aim to enhance the coherence and interoperability of future TCP gene research in plant and crop science. In our view, this work will serve as a valuable resource for researchers studying TCP genes in apple and will provide insights into the evolutionary dynamics and functional roles of TCP genes in plants.DOI:
https://doi.org/10.23796/LJ/2024.008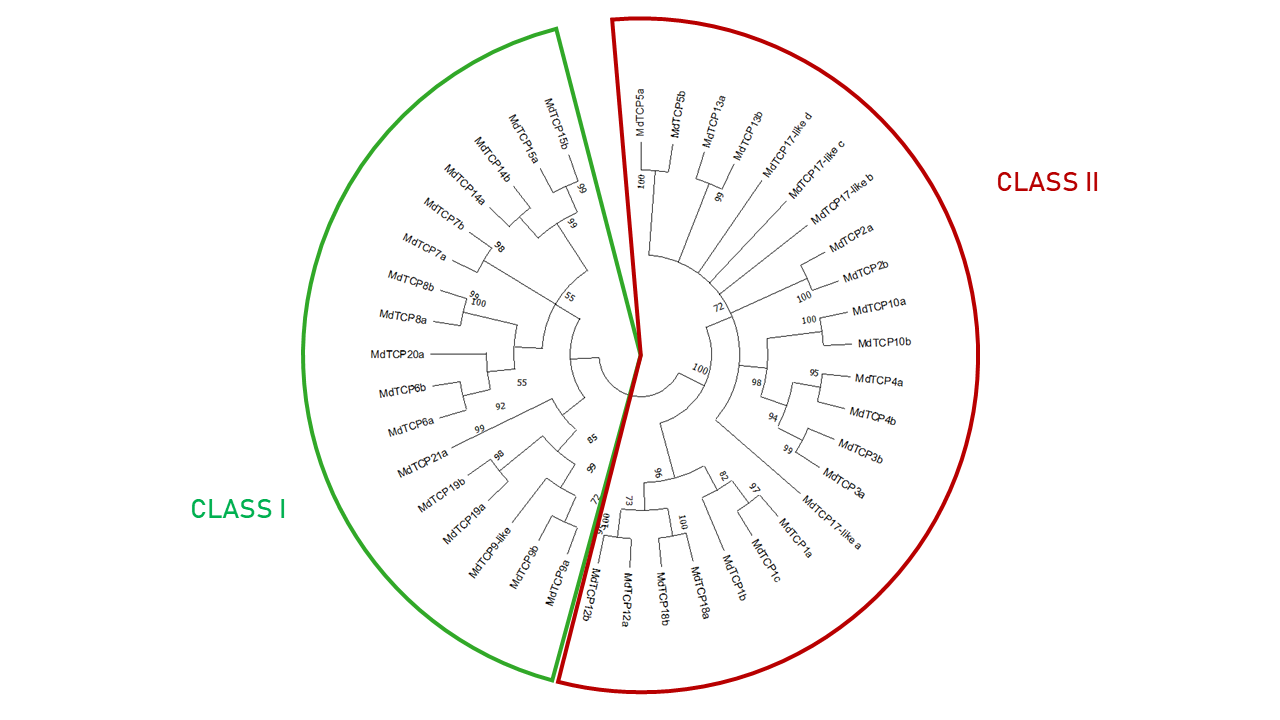
Published
05.04.2024
How to Cite
Tabarelli, M. (2024). From Chaos to Clarity: The Apple TCP Genes Naming Makeover. Laimburg Journal, 6. https://doi.org/10.23796/LJ/2024.008
Issue
Section
Short Papers